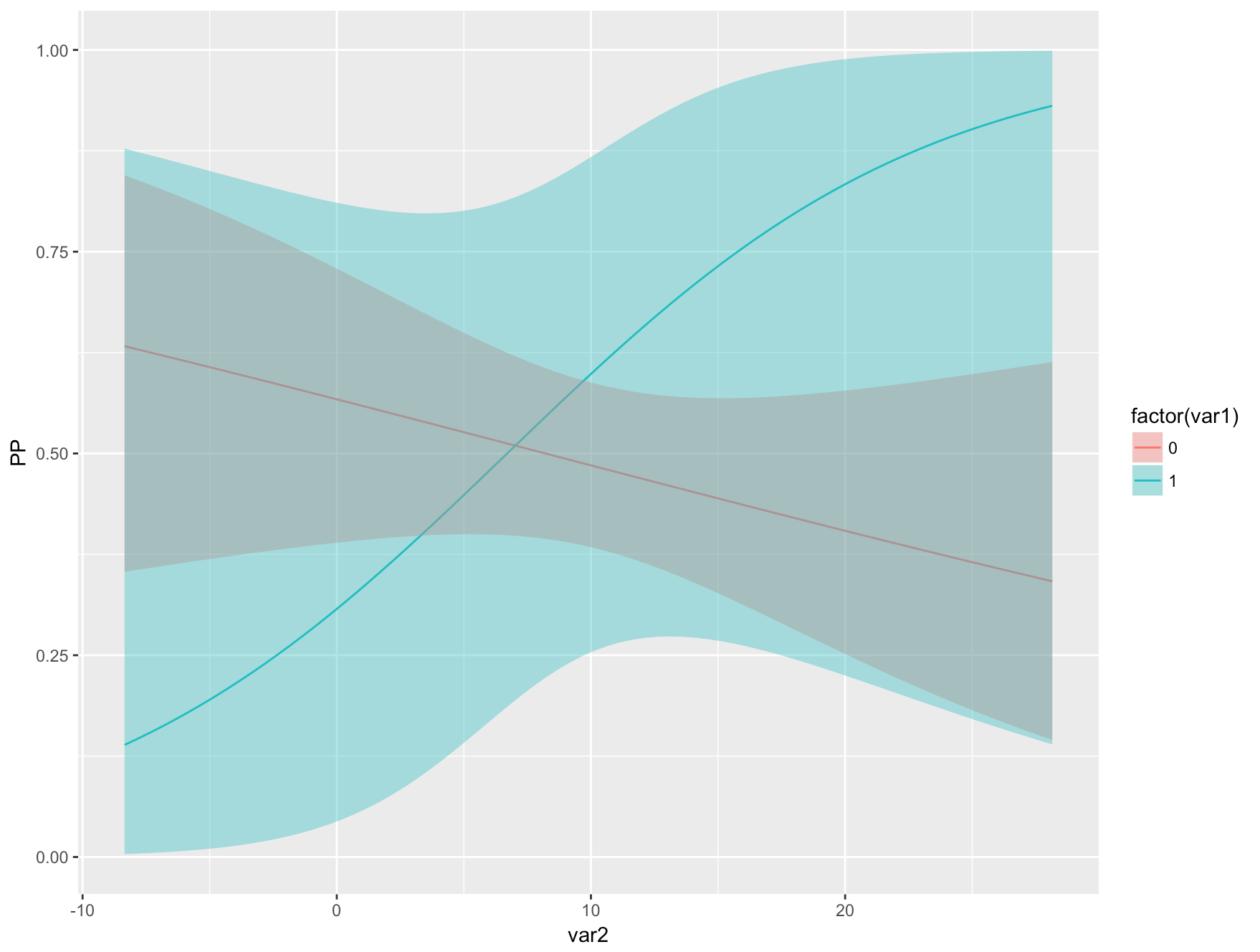
Cplot With Factors R
In R, there is a special data type for ordinal data. This type is called ordered factors and is an extension of factors that you’re already familiar with. To create an ordered factor in R, you have two options: Use the factor function with the argument ordered=TRUE. Use the ordered function. Say you want to. Factor.col If x is a factor variable in the model, the color to use for the border of the points. Factor.fill If x is a factor variable in the model, the color to use for the fill of the points. Factor.cex If x is a factor variable in the model, the “expansion factor” to use for the point size. Draw one or more conditional effects plots reflecting predictions or marginal effects from a model, conditional on a covariate. Currently methods exist for “lm”, “glm”, “loess” class models.
Example
Factors are one method to represent categorical variables in R. Given a vector x whose values can be converted to characters using as.character(), the default arguments for factor() and as.factor() assign an integer to each distinct element of the vector as well as a level attribute and a label attribute. Levels are the values x can possibly take and labels can either be the given element or determined by the user.
To example how factors work we will create a factor with default attributes, then custom levels, and then custom levels and labels.
Instances can arise where the user knows the number of possible values a factor can take on is greater than the current values in the vector. For this we assign the levels ourselves in factor().
For style purposes the user may wish to assign labels to each level. By default, labels are the character representation of the levels. Here we assign labels for each of the possible levels in the factor.
Normally, factors can only be compared using and != and if the factors have the same levels. The following comparison of factors fails even though they appear equal because the factors have different factor levels.
This makes sense as the extra levels in the RHS mean that R does not have enough information about each factor to compare them in a meaningful way.
The operators <, <=, > and >= are only usable for ordered factors. These can represent categorical values which still have a linear order. An ordered factor can be created by providing the ordered = TRUE argument to the factor function or just using the ordered function.

For more information, see the Factor documentation.
6.1 About this chapter
- Questions:
- How can I make plots that compare multiple categories?'
- Objectives:
- Understand factors
- Understand colouring and faceting on factors
- Use factors for summaries and plot design
- Keypoints:
- A factor is a value of a categorical variable, or the different values a label can take
- Factors are needed to subset and add attributes to data dynamically

6.2 Factors
In previous plots we’ve been using categories, specifically the Species category to split our data, colour our plots etc. These categorical columns are called Factors in R. Looking at the diamonds data set we can see how this is set up in R.
Here we can see the cut, color and clarity columns are all non-numeric, textual data. These are the factor variables of this dataset. We can confirm that by asking for the class of the column, that is, the type of data in it. We use the dataset $ column name syntax for this.
We can also ask for all the different values of the factor, in R called the levels
6.3 Colouring by factors
Let’s look at applying mappings by a factor. Let’s look at how price varies by cut.
Now let’s throw a second variable in there, lets see how color varies within each cut. We do this by creating a new aesthetic mapping within the geom_jitter()
The spots are all overlapping, we can force the different colours to stay separate with the position option. We use position_dodge() to make them dodge each other. The width option tells the spots how far to stay apart.
We can also throw other geoms on top in the same way. EG Boxplots for each cut and colour
Remember layers/geoms are independent, so can be set up to show individual aspects of the data. Let’s have a boxplot for the whole of the cut, irrespective of the colour.
And of course, the whole thing still works even if we are comparing two numerical columns. We can still use the aesthetic mapping in the geom to colour our points by a factor
6.4 Small multiple plots
Sometimes, trying to squeeze a lot of data into one plot isn’t the clearest way to show it. Instead small multiple plots (different data, same settings) can be used. In ggplot, this is called faceting and is done with the facet_wrap() or facet_grid() function. We use the factors to define the facet. Let’s add faceting to the previous plot
Plot With Factors In R
Here we see the plot is divided into panels, one for each ‘cut’. The facet_wrap() function puts all the panels into a single row, but wll wrap that row as space demands. The syntax is a bit odd, we used the ~ operator to mean ‘varies by’ , even though we only used one variable. It’s just a quirk of ggplot.
The facet_grid() function forces a grid structure and can take more than one factor. Now the ~ ‘varies by’ syntax makes more sense:
6.5 Quiz
The built in dataset CO2 describes measurement of CO2 uptake versus concentration for Quebec and Mississippi grasses in chilled and nonchilled tests. The dataset is as follows:
Typeis a factor column with two levelsQuebecandMississippiTreatmentis a factor colum with two levelsnonchilledandchilledUptakeis a numerical colum with CO2 uptake rate in micromoles per metre squared per secondPlantis a factor with twelve levels, one for each individual plant assayed.
Cplot With Factors R
- Create a plot with
geom_point()that shows the Plant on the x-axis and the Uptake on the y-axis. Colour the points by ‘Type’ andfacet_wrap()byTreatmentto get a subplot for chilled and nonchilled.